Abstract
Human teeth are biological structures that resist extreme conditions thus becoming a useful source of DNA for human forensic identification purposes. When it is possible, forensic prefer only non-damaged teeth whereas those with cavities are usually rejected to avoid both external and internal bacterial contamination. Cavities are one of the most prevalent dental pathology and its incidence increases with ageing. The aim of this study was to validate the use of teeth with cavities for forensic identification. A total of 120 individual teeth from unrelated patients (60 healthy and 60 with cavities, respectively) extracted by a dentist as part of the normal process of treatment, were submitted for further analysis. Dental pulp was obtained after tooth fragmentation, complete DNA was extracted and the corresponding human identification profile was obtained by the AmpFlSTR NGM SElectTM kit. Cariogenic microbiota was determined by PCR-DGGE with bacterial universal primers and bands were excised, re-amplified and sequenced. From the 120 dental pieces analyzed, a defined genetic profile was obtained in 81 (67.5%) of them, with no statistical differences between the healthy and the cavities-affected teeth. Statistical association between teeth status, DNA content and genetic profiles was not observed. Complex bacterial communities were only detected in the cavities group, being the Streptococcus/Enterococcus, and Lactobacillus genera the most represented. We conclude that teeth with cavities are as valid as healthy dental pieces for forensic human identification. Moreover, the severity of the cariogenic lesion as well as associated bacterial communities seems not to influence the establishment of human dental profiles.
1.Introduction
Teeth are structures that resist very extreme situations such as traumas, incineration, or prolonged expositions to adverse environmental conditions, and due to these attributes, they are commonly used in forensic investigations for human identification purposes. Additionally to the morphological analysis, teeth are an important source of viable human tissue for DNA extraction. The recent molecular tools based on DNA amplification and nucleotide sequence analysis have revolutionized the Forensic Dentistry
allowing exact identifications in missing persons, mass disasters, terroristic acts or crimes scenes.
Dental pulp tissue is usually a suitable material for DNA extraction and its forensic use has been described and validated previously, moreover, different methods to recover the pulp material from the teeth are available [5]. In EU countries it is mandatory to perform the individual genetic identification using this tissue by allele distribution of 15 short tandem repeat (STR) loci previously standardized by the European Standard Set (ESS).
Healthy dental pieces are employed for human forensic identification, whereas those teeth with cavities are usually rejected because their integrity might be compromised and external DNA contamination from different sources cannot be ruled out, thus misleading final results. In absence of healthy pieces, forensic use teeth with cavities, though their real potential has been scarcely explored.
Other important factor to take into account is the bacterial deleterious activity on various human structures. Oral microbiota has an enormous potentiality conditioning health and disease. Classically, Streptococcus mutans has been implicated in the cariogenic pathology, although the recent metagenomic tools have demonstrated the existence of a complex microbiota with different genera and species involved in this issue. The influence of this cariogenic microbiota on DNA extraction from human teeth has not been yet evaluated. The aim of this work was to assess the utility of DNA obtained from teeth with cavities in comparison with healthy teeth for human forensic identification.
2. Material and Methods
2.1 Teeth collection
During 2011–2012, a total of 120 teeth’s structures (adaman- tine, cameral pulp and root) were collected, 60 pieces from healthy subjects and 60 with different degree of cavities affectation. In the case of healthy individuals, teeth were extracted in the course of orthodontic or periodontal diseases treatments. Each patient signed a consent that has been previously approved by the Ethics Committee of the University Alfonso X El Sabio, Madrid. The inclusion criteria for the group of patients with cavities were the presence of this pathology diagnosed either visually or by radiographic techniques, whereas teeth with external or internal root resorption, open root apex, fenestrations in alveolar bone, endodontic teeth, root fractures, etc. were rejected.
2.2. DNA extraction
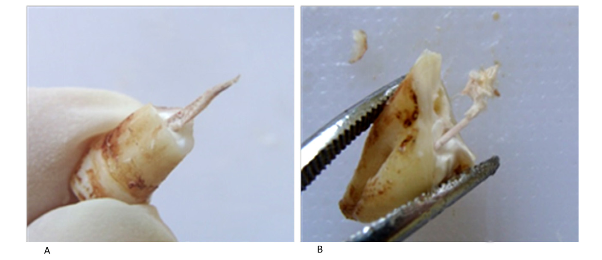
After the extraction, each dental piece was conserved in sterile gauze at 4 8C until its subsequent processing. A mechanical manual and fast fragmentation in one hit was applied to each piece, collecting the pulp tissue from the pulp chamber and the root canal without pollutants (Fig. 1). Pulp tissue was collecting in an eppendorf tube with 1 ml of saline and conserved refrigerated until DNA extraction.
DNA from the total pulp material was extracted by the commercial QIAamp1 Mini Kit (Qiagen, Hilden, Germany) following the manufacturer instructions for tissues. Human DNA concentration was determined by the QuantifilerTM System Human DNA Quantification Kit (Life Technologies, Foster City, CA, USA), whereas NanoDropTM Lite Spectrophotometer (Thermo- Scientific) was used to determine the total concentration and the purity of the samples through the A260/280 coefficient.
2.3. Human genetic profile
The genetic profile of each subject was obtained using the previously approved European standard Set STR or ESS type markers (Council resolution (EC) of 30 November 2009). Genomic DNA was amplified using the AmpFlSTR1 NGM SElectTM (Life Technologies, Foster City, CA, USA) kit according to the manufacturer’s instructions in a final PCR volume of 25 ml using as template 1–1.5 ng of DNA from each sample. Amplicons were analyzed by capillary electro- phoresis in an automatic sequencer ABI-PRISM-3130 (Life Technol- ogies, Foster City, CA, USA). Analysis of results was performed by the ‘‘GeneMapper’’ software program (versions ID v3.2 and ID-X), obtaining four different possibilities: (1) good profile (10 STR markers autosomal and amelogenin gene); (2) partial profile (5–9 STR markers autosomal and the amelogenin gene), (3) low profile (5 STR markers autosomal and the amelogenin gene), and (4) no profile, impossibility of identification.
2.4. Cariogenic bacterial microbiota
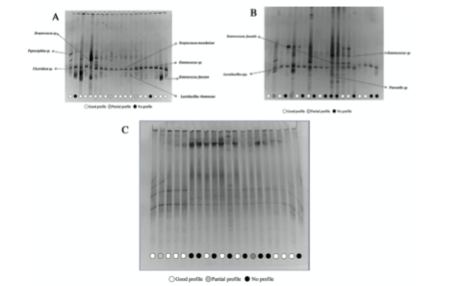
he bacterial microbiota associated with the 120 teeth was analyzed by the denaturing gradient gel electrophoresis (PCR- DGGE) technique, using primers and conditions previously published. Positive 16S-rDNA amplicons were separated in 8% vertical electrophoresis polyacrylamide gels at 60 8C; with a ureaformamide denaturating gel gradient of 30–50% at 130 V during 16 h. Bands were excised, re-amplified with the same primers, sequenced and results were compared with those from the GenBank NCBI database to assign the corresponding genera and species.
2.5. Statistical analysis
Data are presented as median and interquartile range (Percentiles 25 and 75). Chi-square test has been used to compare the proportion of obtained genetic profiles between both samples, healthy and cavities tooth. The U of Mann-Whitney test was applied to compare the concentration and the purity of both groups, whereas Fisher test was used to compare the grade of human identification profile. Statistical analysis was performed with the Stata v.13 (StataCorp. 2013. Stata Statistical Software: Release 13. College Station, TX: StataCorp LP) software. Only p-values of ≤ 0.05 have been considered statistically significant.
3. Results
All subjects included in this study were from the same geographical area and the same dentist performed dental extractions. Gender distribution was: 23 males/37 females for
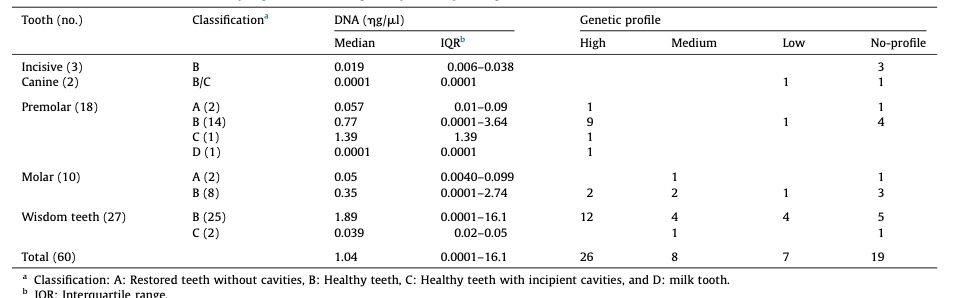
the control group, and 35 males/25 females for the group affected with cavities. Age ranges were as follows: 0–29 years (27 subjects in the control group/8 in the group with cavities); 30–59 years (24/ 42), and 60–99 years (9/9). Further data from the subjects included in the study and features from dental pieces of both groups are detailed in Table 1 and Table 2.
The fragmentation method allowed the complete extraction of the pulp tissue in most of the teeth (Fig. 1), although scraping the camera was necessary in those pieces with advanced cavities. DNA extraction was successful in both groups, obtaining a variable range of total concentration whereas purity values were homogeneous (Fig. 2).
From the 120 dental pieces analyzed, a genetic profile was obtained in 81 of them (67.5%), without statistical differences in both groups, 41/60 (68.3%) in the control group and 40/60 (66.6%) in the group with cavities (p = 0.846) (Tables 1 and 2). Neverthe- less, the distribution between full, partial and low profiles was higher among the cavities group (35/3/3) than in the control group (25/9/7). No statistical association between tooth type, DNA concentration or purity and genetic profiles was obtained.
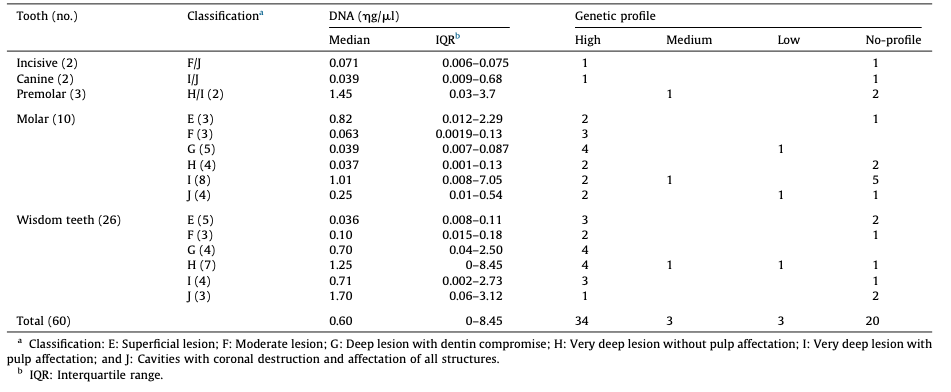
Bacterial DNA amplification was not obtained in any of the 60 pieces from the control group, whereas a complex bacterial profile was obtained in 40 out of the 60 samples in the cavities group. In general, complex bacterial communities were detected within this latter group, being Streptococcus/Enterococcus and Lactobacillus the most represented genera (Fig. 3). The statistical analysis failed to establish an association between the microbiota and the human profiles.
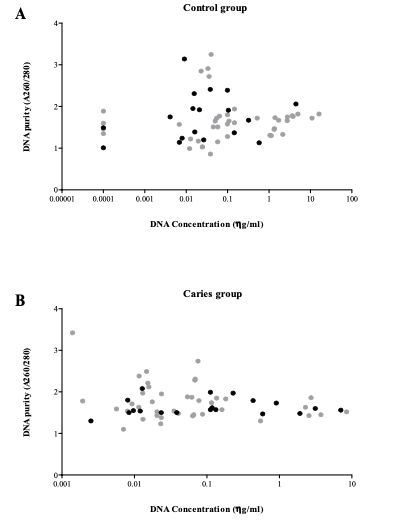
Grey points represent those cases in which a human profile could be achieved, whereas
black points represent those cases in which no identification was obtained.
Several methodologies for tooth sampling and DNA extraction have been validated, being the type of processing a key determinant of the quality of the sample. In this work we decided not to decontaminate teeth because this treatment might decrease the efficiency of the DNA extraction, as previously reported, moreover, teeth were extracted from living persons and dental pieces were conserved. On the contrary, teeth exposed to adverse environmental conditions might be decontaminated. Negative results obtained in DGGE experiments from healthy teeth confirmed that previous manipulation was adequate, and pieces were adequately preserved from external contamination.
The way to accede to the pulp camera is also an important decision thus we chose the manual fragmentation instead other methods that might decrease the quality and quantity of the DNA extraction. In our experience, fragmentation is an easy technique that retrieves a high recovering of pulp tissue.
As stated, all subjects included in this study belong to the same geographical area and differences between gender and age were matched with their dental pathology. The control group had underlying orthodontic causes for dental extractions, and as they were younger they presented a higher proportion of premolar teeth than the subjects of the other group. Interestingly, a premolar milk tooth was included through which a full identification profile could be obtained. This fact has been published also by other authors.
The concentration and the purity of the DNA samples are limiting factors to obtain satisfactory results in human forensic identification. For purity, 1.8–2.0 values are the most adequate, less than 1.8 indicates protein or membranes contamination, and higher to 2.0 point to the presence of mineral salts or RNA. In our collection, purity was adequate and uniform for all samples, whereas a wide range of DNA concentration was observed. Unexpectedly, a good human profile was obtained even from samples with 0.001 ng/ml of DNA. In the case of human profile and bacterial microbiota determinations, no results were obtained in approximately a third of the samples, even though amplification experiments were repeated in independent days. A possible explanation for this relies in that both techniques are based on PCR amplification processes, which might be very much affected and limited by the samples’ conditions. Other authors have reported similar results in their series.
5. Conclusion
Achievement of human profile in the cavities group was identical to that of the control group, thus validating the use of teeth with cavities for human forensic identification. The severity of the cariogenic lesion as well as the associated bacterial communities seem to have not influence in the human profile identification protocol. Statistical analysis demonstrated that in our series, successful results in human profile determination were not associated with either DNA concentration or its purity values.
Acknowledgements
We thank Dr. Beatriz Romero for scientific advice and to Dr. Marı ́a Isabel Morosini for critical review of the manuscript. This work was partially supported by the PI13/205 project from the Instituto de Salud Carlos III (Ministry of Economy and Competi- tiveness) and co-financed by the European Development Regional Fund ‘‘A Way to Achieve Europe,’’ ERDF, Spanish Network for the Research in Infectious Diseases (REIPI RD12/0015).